VIAMD
Visual interactive analysis of molecular dynamics
What is VIAMD
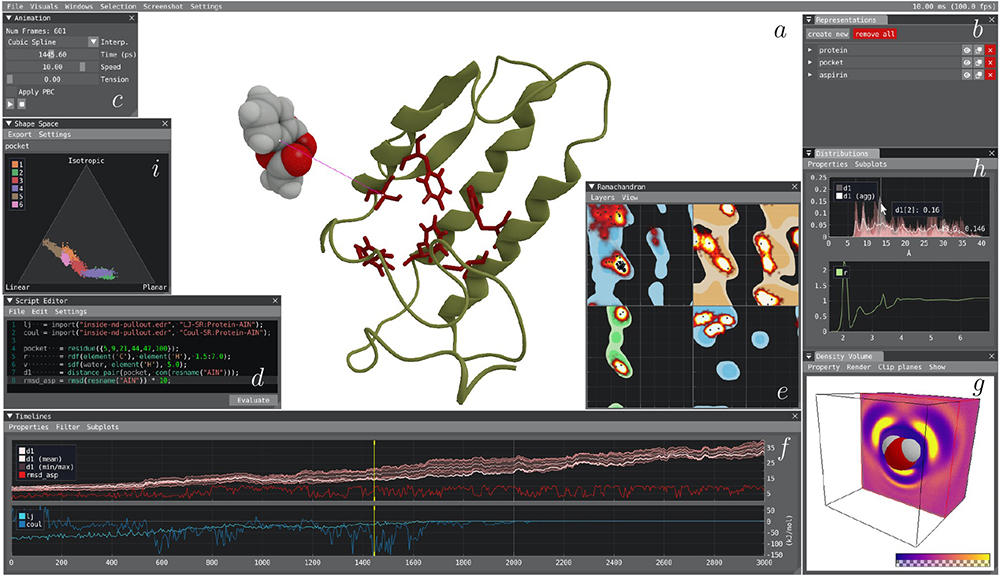
VIAMD is an interactive analysis tool for molecular dynamics (MD) written in C/C++. It is designed to facilitate the exploration of large-scale spatio-temporal molecular dynamics simulation data. It enables users to visually analyse and explore molecular dynamics simulations by offering an interactive scripting interface. This interface allows users to define and evaluate properties of interest. The application also provides context-aware suggestions and feedback on expressions, including type information and visualizations. By utilising the various components of VIAMD, users can explore and analyse user-defined properties, allowing for correlation with spatial conformations, statistical analysis of distributions, and powerful aggregation of multidimensional properties such as Spatial Distribution Functions.
VIAMD development partners
VIAMD is developed by researchers from the Department for Media and Information Technology at Linköping University, Norrköping, and from the PDC Center for High Performance Computing at the KTH Royal Institute of Technology, Stockholm.
Current developments
The VIAMD team is currently working on adding support for VeloxChem files to VIAMD in order to efficiently evaluate and render molecular orbitals under different modalities, making it possible, for instance, to undertake interactive analysis of absorption spectra of large molecular systems in terms of natural transition orbitals.
Links
- Paper introducing VIAMD code: doi.org/10.1021/acs.jcim.3c01033
- VIAMD code on GitHub: github.com/scanberg/viamd
- Tutorial videos about VIAMD on YouTube: bit.ly/4aRsPrh
- Webinars: bioexcel.eu/webinar-viamd-a-software-for-visual-analysis-of-molecular-dynamics-2024-02-06
- VIAMD on Twitter (@VIAMD_): twitter.com/VIAMD_
Contact
For more information about VIAMD development at PDC, you are welcome to contact:
