Update on BioExcel and PerMed CoEs
Alessandra Villa and Rossen Apostolov, PDC
BioExcel Centre of Excellence
BioExcel@PDC is continuing its broad range of initiatives that support the computational biomolecular research community.
BioExcel Webinar Series
The BioExcel webinar series ( bioexcel.eu/category/webinar ) continues to feature notable developments in the field of computational biomolecular research. For example, in February, Paul Bauer (from the Department of Applied Physics at the KTH Royal Institute of Technology) introduced the features that are new in GROMACS for 2022 ( youtu.be/yIblJjez6P0 ), and in April, Szilárd Páll (from PDC) spoke about GROMACS’ performance, flexibility and portability and how advances in these areas can be applied to real-world simulations ( youtu.be/rTUz28f8p6g ). If you missed any of the webinars, you can catch up on the videos on BioExcel’s YouTube channel: www.youtube.com/playlist?list=PLzLqYW5ci-2fanl2RtYHyFvjftu_M4Jwy . Remember to also keep an eye on www.bioexcel.eu/webinars for upcoming events!
Online Biomolecular Simulation Workshops
Online biomolecular simulation workshops were held in spring, both at the basic and advanced levels (see bioexcel.eu/news-and-events/past-events ). The “physical” (rather than online) BioExcel Summer School will be run again in June this year after a break of two years. For details, see: bioexcel.eu/events/bioexcel-summer-school-on-biomolecular-simulations-2022 .
Biomolecular Simulation Software Use Cases
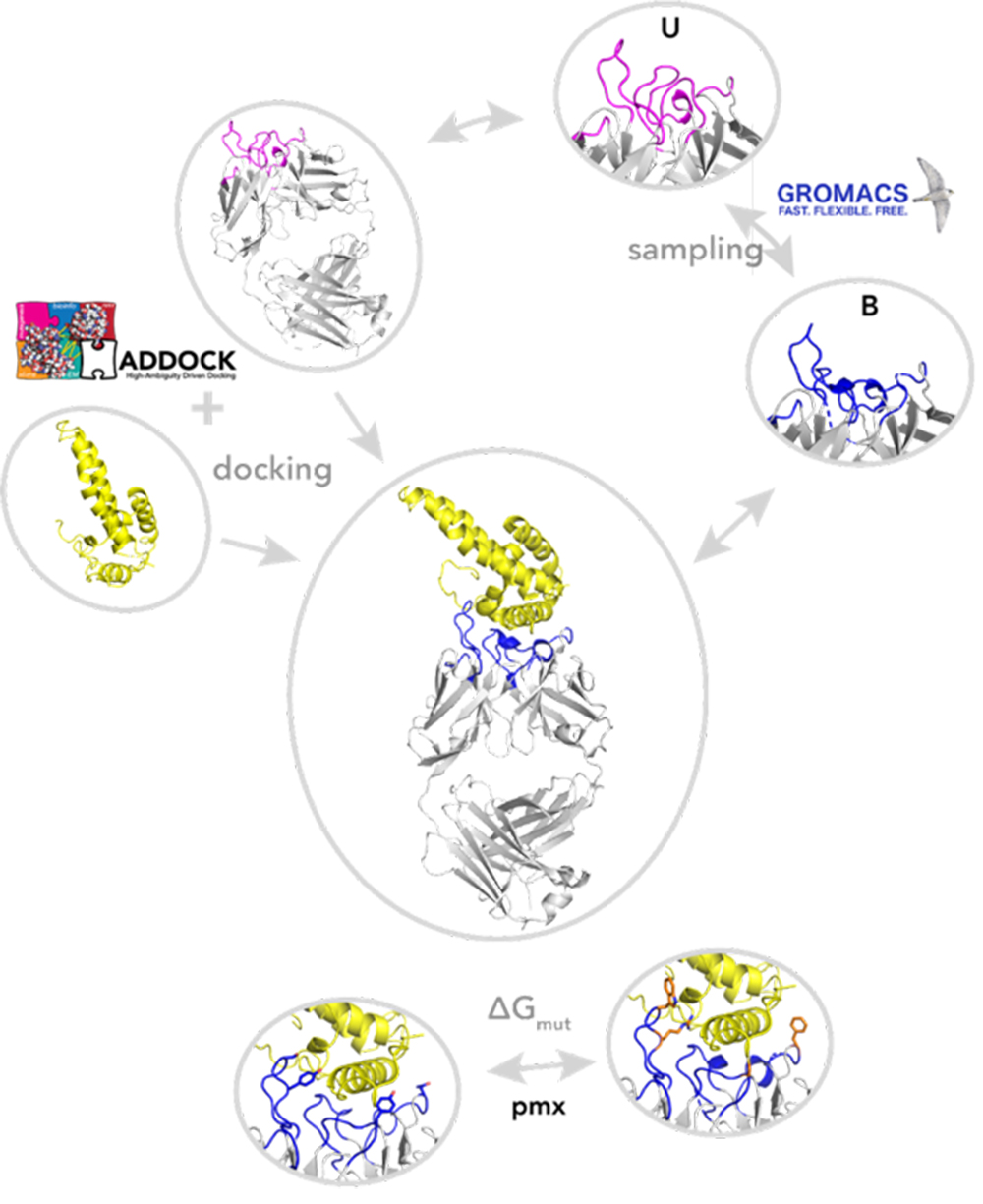
BioExcel has recently published several use cases involving different types of biomolecular simulation software. These include “Electronic Interaction Phenomena: Proton Dynamics And Fluorescent Proteins”, which looks at using the GROMACS, CP2K, CPMD, MiMiC and pmx software and can be found at www.hpccoe.eu/2022/03/21/electronic-interaction-phenomena-proton-dynamics-and-fluorescent-proteins , and “Molecular dynamics meet docking: A use case on antibody design”, which features HADDOCK and GROMACS (see www.youtube.com/watch?v=_TDKfKX4kwM ).
For everyone who is interested in learning more about how to use GROMACS, BioExcel is continuously updating the tutorials at tutorials.gromacs.org . The latest tutorial is on alchemical transformation using the AWH method ( tutorials.gromacs.org/awh-free-energy-of-solvation.html ). If you would like some tips on using social media for communicating about science, check out Michelle Mendoca’s webinar: bioexcel.eu/webinar-social-media-in-science-communication-2022-02-17 . Anyone who is active in the area of biomolecular simulations is welcome to join the BioExcel community and participate in the active BioExcel forums (see ask.bioexcel.eu and also gromacs.bioexcel.eu ).
PerMed Centre of Excellence
PerMed@PDC has been contributing to developing a competency hub in computational personalised medicine ( competency.ebi.ac.uk/framework/permedcoe/2.0 ). In general, this Competency Hub (see competency.ebi.ac.uk ) is a website where you can browse competencies, career profiles and training resources in the bio-related computational areas to help advance your career. For example, there is a section for professionals in computational personalised medicine. PerMedCoE also recently developed two learning pathways (one based on the Linux command line and the other using Jupyter Notebook) that can help the biomedical community to use high-performance computing (HPC) infrastructures more easily.
Advanced GROMACS Workshop
BioExcel, in collaboration with the CSC – IT Center for Science, Finland, and the EuroCC National Competence Centre Sweden (ENCCS), held an online GROMACS workshop on 7-9 February this year. The workshop was supported by EuroCC and covered various topics that improve efficiency when using GROMACS: enhanced sampling techniques by Berk Hess and Alessandra Villa (from KTH), QM/MM with CP2K by Dmitry Morozov (from the University of Jyväskylä, Finland), performance optimisation for CPU and GPU by Artem Zhmurov (from ENCCS), automation via BioBB-workflows by Adam Hospital (from the Institute for Research in BioMedicine, Barcelona) and GROMACS Python-API by Eric Irrgang (from the University of Virginia, USA). The workshop targeted advanced GROMACS users. To avoid problems with inexperienced attendees, we provided introduction material on the workshop topics (which is available online at ssl.eventilla.com/advanced-gromacs-2022 ). The workshop was run using an open on-demand interface that provided the attendees and trainers with easy access to the Jupyter Notebook environments and to the HPC resources. It was the first time that CSC used this setting and, apart from some small technical problems, overall the attendees found the hands-on approach easy to follow and enjoyed being active in the live documents (using the HackMD platform).
